Rolling circle–based linear amplification (RCA) is used to produce single-stranded DNA molecules composed of multiple concatemerized copies of equally represented DNA strands of each particular DNA fragment. The amplification is carried out using an artificial thermostable polymerase having a strong strand displacement activity (9). This allows multiple cycles of denaturation-annealing-extension to ensure efficient and less biased amplification in a reaction we termed pulse-RCA. Because all these copies are independent replicas of the original DNA fragment, potential errors of amplification remain unique for each copy and do not propagate further. Copies of opposite strands are in an end-toend orientation and separated by common spacers used as PCR priming sites during the second step of the process when concatemerized copies are individually amplified and converted into a sequencing library. Thus, the resulting sequencing library is composed of PCR duplicates of multiple independent copies of an original DNA fragment assembled in rolling circle (RC) amplicons.
SCIENCE
Somatic mutations and structural variants have been found associated with human disease, including cancer as well as the aging process itself.
Most information on somatic mutations has come from studying clonally amplified mutant cells, based on a growth advantage or genetic drift. However, almost all somatic mutations are unique for each cell, and the quantitative analysis of these low-abundance mutations in normal tissues remains a major challenge in biology.
Our novel approach can accurately identify point mutations and DNA structural variants in normal cells and tissues.
Rolling Circle Amplification
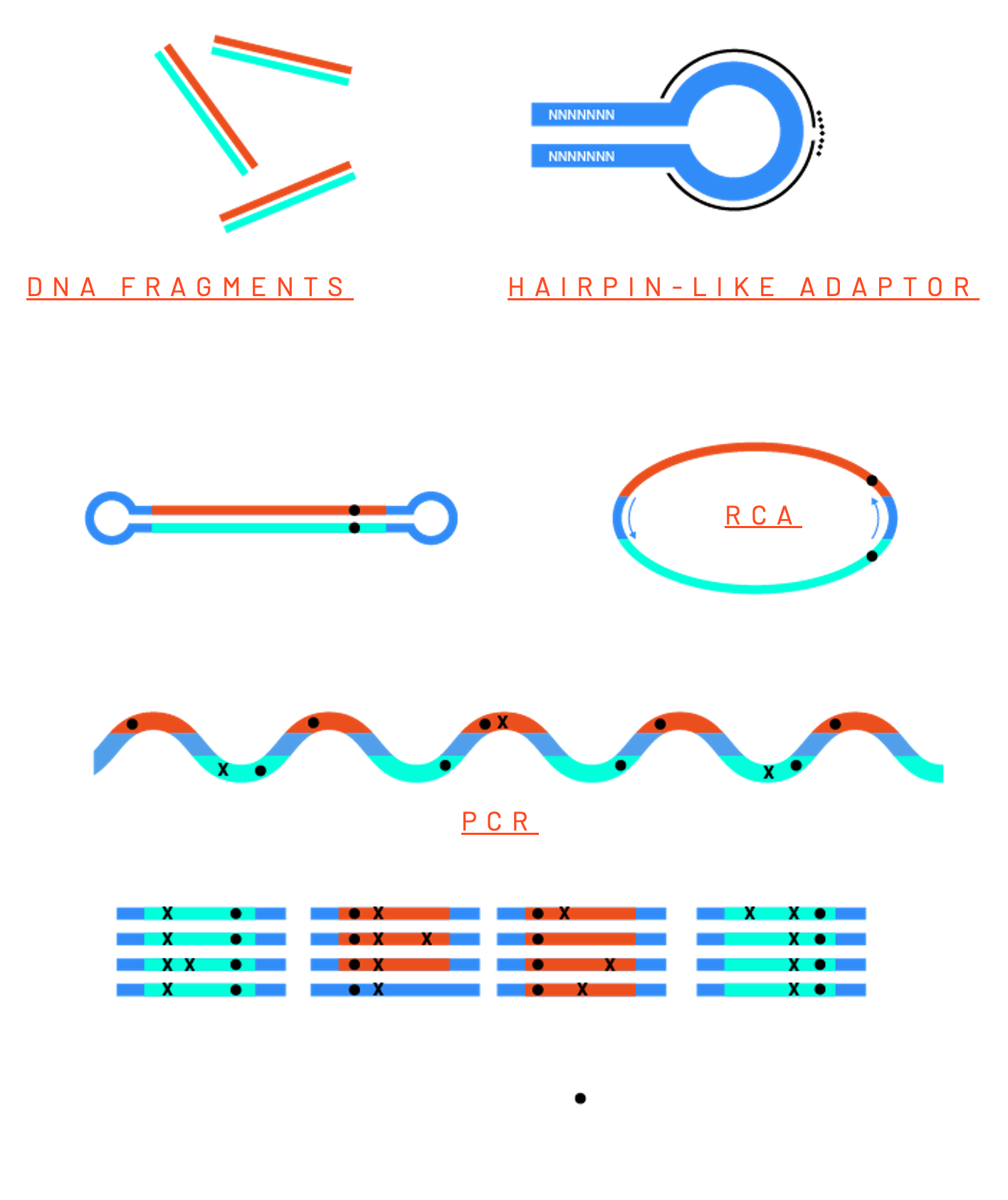
Removing false positives
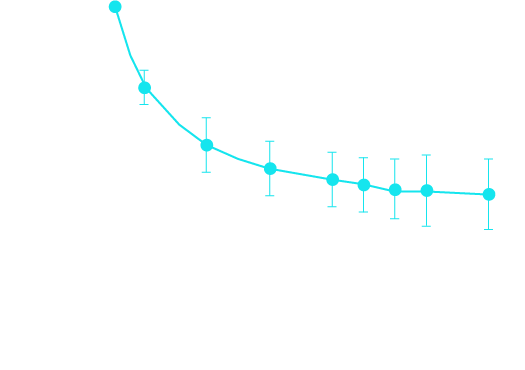
We found that increasing the minimum required strand family size from two to seven led to a statistically significant decrease in observed mutation frequency at each iteration, resulting in a more than twofold decrease (54% change).
SMM-Seq vs SC-seq
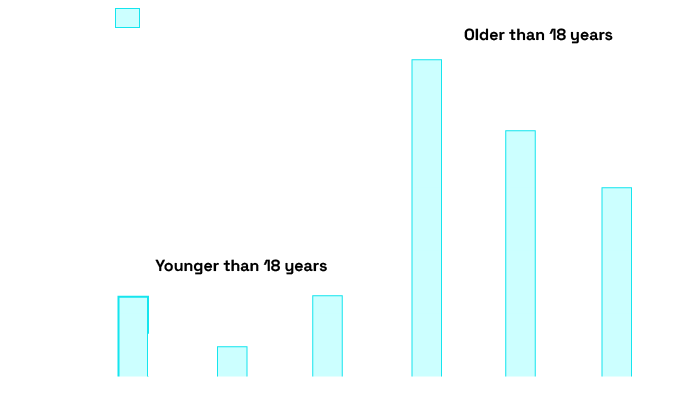
Comparison between SMM-Seq and single cell (SC-seq) shows accurate concordance between techniques.
SNVs accumulate during aging
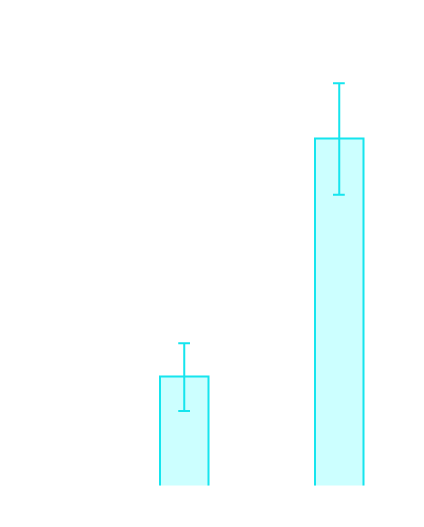
SMM-seq confirmed the age-related elevation in the somatic mutation frequency
References
The work presented above has been recently published and can be found here:
Maslov, A. Y., Makhortov, S., Sun, S., Heid, J., Dong, X., Lee, M., & Vijg, J. (2022). Single-molecule, quantitative detection of low-abundance somatic mutations by high-throughput sequencing. Science Advances, 8(14), eabm3259